RESEARCH
Glyco-Systems biology Division
Purpose / Contents

Deciphering glycan code from glycan big data
The purpose of the Systems Biology Division in the Integrated Glyco-Big Data Center (iGDATA) is to understand glycan functions and complexity by analyzing glycan-related big data using systems biology approaches. This will lead to the prediction of key biomarkers and drug targets. By collaborating with other divisions focusing on studies using model organisms and human samples, we will predict glycan structures associated with various diseases. To achieve this, we will develop algorithms, tools, and software targeting mass-spec-based glycomics and glycoproteomics data. In addition, we aim to develop a new interface that links genome science and glycoscience, which is an innovative platform for developing new strategies against various diseases.
Examples
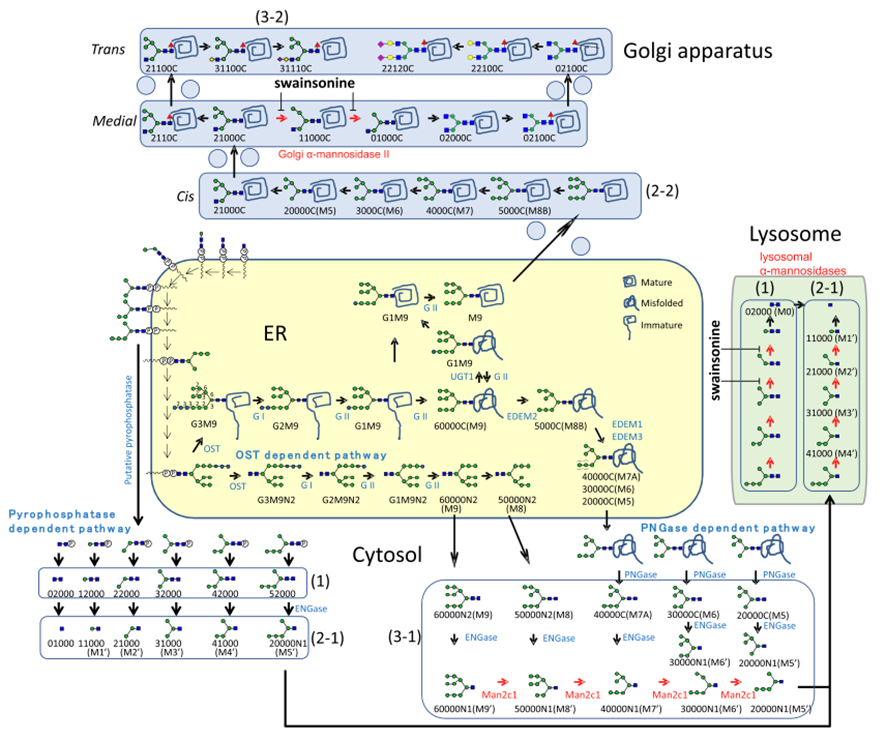
Comprehensive understanding of changes in cellular glycans using mass-spectrometry
We have developed a new comprehensive method for the glycomic analysis of total cellular glycans using mass-spec, which enables us to detect glycan changes in various samples. Recently, we elucidated the changes in glycan structure in cells caused by the plant compound "swainsonine“, This compound is known to evoke toxic symptoms in domestic animals.
(Morikawa et al., BBA - General Subjects, 2022, 1866, 130168)
Members List
Yusuke Matsui
Division headSystems Biology Division
- Research interests
- Statistical science, informatics, computational biology, computational neuroscience, bioinformatics
- Research subject
- Life science is generating huge and complex data at an unprecedented rate. Various and heterogeneous big data are being accumulated, including next-generation sequencing technology, mass spectrometry to capture proteome and post-translational modifications, and sensing technology to capture imaging and biological information. It is quite important to utilize such large-scale big data in life science in order to reveal the mechanisms of unknown biological systems. Our mission is to develop useful mathematical modeling and data analysis methods in life science.
Motonori Ota
Systems Biology Division
- Research interests
- Structure bioinformatics, protein tertiary structure, naturally denatured protein, protein-protein interaction network
- Research subject
- Our research focuses on bioinformatics related to protein conformations and how interactions and conformational changes lead to functions. We also develop algorithms (methods) for data analysis and constructing databases.
Naoki Honda
Systems Biology Division
- Research interests
- Data-Driven Biology, Machine Learning, Mathematical Modeling
- Research subject
- By making full use of machine learning, we are developing mathematical modeling research rooted in data. We are also conducting single-cell omics analysis and pathological analysis.
Kensaku Mori
Systems Biology Division
Morten Thaysen-Andersen
Systems Biology Division
- Research interests
- Clinical glycoproteomics, Glycoimmunology, N-glycosylation, Cancer, Sepsis, Innate immunity
- Research subject
- The Glycoproteomics Lab@iGCORE develops and applies cutting-edge LC-MS/MS-based methods for quantitative and comparative glycoproteomics of human biospecimens to holistically explore elusive roles of protein N-glycosylation in human glycobiology with a particular focus on the innate immune system. The group uses high throughput glycoproteomics methods compatible with large clinical sample cohorts to study how the N-glycoproteome is remodelled with aberrant physiology and with various disorders including cancer, inflammation and infectious diseases.
Hiroyuki Kaji
Systems Biology Division
- Research interests
- Glycoproteomics, glycome, proteome, liquid chromatography, mass spectrometry
- Research subject
- We develop and apply techniques for comprehensive analysis of post-translational modifications of proteins, especially glycosylation, using mass spectrometry. We systematically analyze the structures of glycans attached to the glycosylation sites on each glycoprotein in biological samples such as body fluids, cells, and tissues, and when and how these glycans change. By obtaining this information, we hope to contribute to basic research such as elucidation of the involvement of glycans in biological phenomena as well as applied research on development of diagnostic markers and therapeutic targets presenting the altered glycans.
Jennifer Jean Kohler
Systems Biology Division
- Research interests
- chemical biology, fucose, glycolipids, intestinal epithelia, mucus, infectious disease, genetic disorders of glycosylation
- Research subject
- The complex structures and properties of glycans are critical to their myriad biological functions. However, this complexity leads to technical challenges. To tackle these challenges, our research team has created chemical biology methods aimed at understanding glycan function. In particular, we developed photocrosslinking sugar analogs that can be metabolically incorporated into cellular glycoconjugates and used to covalently capture transient glycan-mediated interactions. Using one of these photocrosslinking sugars, we discovered that cholera toxin recognizes fucosylated glycoconjugates displayed on the surface of human intestinal epithelial cells. In current work, we are continuing to develop and apply chemical biology tools to problems in glycoscience. Additionally, we are probing the mechanisms that regulate production of glycoconjugates that comprise the mucosal layer of the intestinal epithelial. Our studies have relevance to infectious disease, cancer biology, and genetic disorders of glycosylation.
Jun-ichi Furukawa
Systems Biology Division
- Research interests
- Glycomics, glycoprotein, glycosphingolipid, glycosaminoglycan, free oligosaccharide, SALSA method
- Research subject
- Cell surface is covered with various glycans whose levels and structures are known to change dramatically with cellular conditions and the environments. Various classes of glycans are present, including complex glycoconjugates such as glycoproteins and glycolipids, glycosaminoglycans such as heparan sulfate and chondroitin sulfate, and free oligosaccharides. We have developed a technique for comprehensive analysis of glycans and conduct total glycomics research on blood, cells, and tissues.
Rebeca Kawahara
Systems Biology Division
- Research interests
- Clinical glycoproteomics, multi-omics, data integration, glycosignatures, diseases
- Research subject
- The focus of my research at the Glycoproteomics Lab@iGCORE consists in developing and applying advanced mass spectrometry-based glycoanalytical methods and multi-omics data integration systems in large cohorts of clinical samples to enable comprehensive and holistic profile of the human glycoproteome and the discovery of new glycosignatures associated with human diseases.
Ryuji Kato
Systems Biology Division
- Research interests
- Image analysis, cell morphology, cell adhesion, cell quality control
- Research subject
- We aim to develop cell image analysis technique for cell quality control and develop culture scaffold materials for controlling cell quality. For this purpose, we label glycolipids in cell membranes and search for glycan binding peptides in cell membranes.
Hisatoshi Hanamatsu
Systems Biology Division
- Research interests
- Glycan analysis, mass spectrometry, sialic acid
- Research subject
- To easily analyze various classes of complex glycoconjugate glycans in cells, tissues, and body fluids, we focus on the development of new subglycome analysis techniques, such as a chemical approach to identify sialic acid linkage patterns by mass spectrometry, a chemical cleavage method for O-type glycans without useful cleavage enzymes, and a new separation method for glycosaminoglycan disaccharides, to elucidate various glycan functions.
Shiori Go
Systems Biology Division
- Research interests
- Glycolipid, glycoproteomics, intracellular trafficking
- Research subject
- We focus on the function of membrane microdomain in various biological phenomena on brain. In membrane microdomain, glycosphingolipids and specific glycoproteins are enriched, interact with each other, and regulate various biological functions. We are particularly interested in the elucidation of comprehensive functions of microdomains glycosylation in neural functions. We analyze structure of glycosphingolipids and glycans of glycoproteins in microdomain using methods for glycoproteomics and glycosphingolipids analysis.
Bingyuan Zhang
Systems Biology Division
- Research interests
- Statistical Science, Machine Learning, Bioinformatics
- Research subject
- Massive amounts of data from innovative technologies such as sequencing, mass spectrometry techniques present new challenges and exciting opportunities. My mission is to develop useful mathematical tools based on state-of-the-art statistical and machine learning techniques to utilize these large-scale heterogeneous real-world data to discover new mechanisms in unknown biological systems and ultimately contribute to scientific discovery.
Akihiro Fujita
Systems Biology Division
Huang Chengcheng
Systems Biology Division
- Research interests
- Chemical glycobiology, Sialic acid, free oligosaccharides, serum
- Research subject
- My work at iGCORE focuses on incorporating chemically labeled glycan analogs into animal cells for in vivo functional studies of glyconjugates. In particular, I use photocrosslinking sialic acid analogs so that the modified glycans can be used to covalently capture transient glycan-mediated interactions. I also aim to develop novel click chemistry method for the fluorescent labeling of sialic acid in animal cells. My research interests also include the structural analysis of free oligosaccharides in serum and their potential biological functions.
Priya DIPTA
Systems Biology Division
Research Division
Contact us by phone

052-789-5365
( +81-52-789-5365 )
Weekday, 9:00-17:00
Contact us




